wcspikes (run-stochastic.xml)
Cable with propagating spikes, varying discretization, stochastic
Total CPU time 24.04 seconds; at 18:18:00 Fri 28 Sep 2007
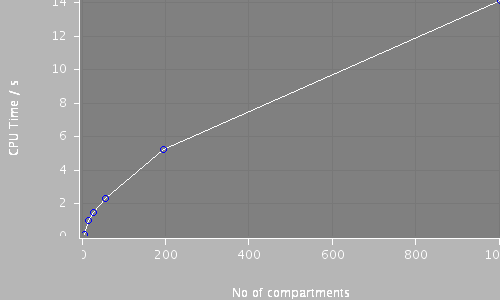
| Compartments | Stochastic channels / cpmts | Continuous channels / cpmts | time/ms | timestep/ms | baseElementSize | CPU Time / s |
| 1002 | 116239 / 1002 | 0 / 0 | 250.0 | 0.05 | 1 | 14.1 |
| 194 | 116238 / 194 | 0 / 0 | 250.0 | 0.05 | 3 | 5.178 |
| 55 | 116238 / 55 | 0 / 0 | 250.0 | 0.05 | 7 | 2.273 |
| 26 | 116239 / 26 | 0 / 0 | 250.0 | 0.05 | 12 | 1.427 |
| 13 | 116239 / 13 | 0 / 0 | 250.0 | 0.05 | 20 | 0.954 |
| 4 | 12566 / 2 | 103672 / 4 | 250.0 | 0.05 | 50 | 0.09 |
 |
Predefined views
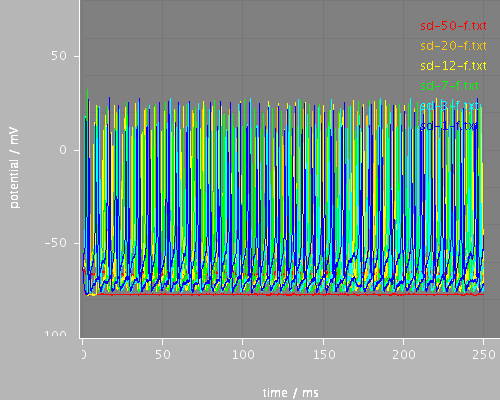
sd-whole
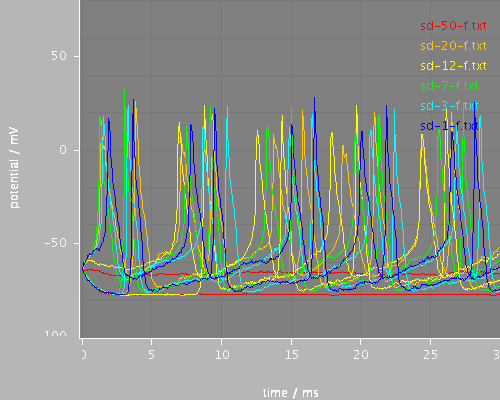
sd-start
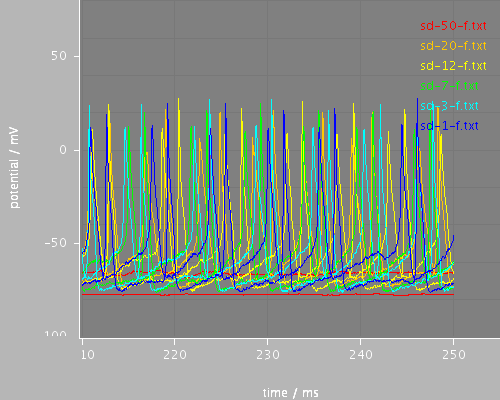
sd-end
All files
Model
Archive file of the complete model: wcspikes.jarrun-stochastic.xml
<PSICSRun timeStep="0.05ms" runTime="250ms" startPotential="-65mV"
morphology="cell"
environment="environment"
properties="membrane"
access="recording"
stochThreshold="10000">
<StructureDiscretization baseElementSize="3um"/>
<info>Cable with propagating spikes, varying discretization, stochastic</info>
<RunSet vary="baseElementSize" values="[1, 3, 7, 12, 20, 50]um"
filepattern="sd-$.txt"/>
<ViewConfig>
<LineGraph width="500" height="400">
<XAxis min="0" max="250" label="time / ms"/>
<YAxis min="-80" max="60" label="potential / mV"/>
<LineSet file="sd-50-f.txt" color="red"/>
<LineSet file="sd-20-f.txt" color="orange"/>
<LineSet file="sd-12-f.txt" color="yellow"/>
<LineSet file="sd-7-f.txt" color="green"/>
<LineSet file="sd-3-f.txt" color="cyan"/>
<LineSet file="sd-1-f.txt" color="blue"/>
<View id="sd-whole" xmin="-10." xmax="260." ymin="-100." ymax="80."/>
<View id="sd-start" xmin="0." xmax="30." ymin="-100." ymax="80."/>
<View id="sd-end" xmin="210." xmax="255." ymin="-100." ymax="80."/>
</LineGraph>
</ViewConfig>
</PSICSRun>
cell.xml
<CellMorphology id="cell"> <Point id="p0" x="0" y="0" z="0" r="0.5"/> <Point parent="p0" id="p1" x="1000" y="0" z = "0" r="0.5"/> </CellMorphology>
membrane.xml
<CellProperties id="membrane"
cytoplasmResistivity="100ohm_cm"
membraneCapacitance="1uF_per_cm2">
<ChannelPopulation channel="na1" density="12per_um2"/>
<ChannelPopulation channel="k1" density="25per_um2"/>
</CellProperties>
environment.xml
<CellEnvironment id="environment"> <Ion id="Na" name="Sodium" reversalPotential="40mV"/> <Ion id="K" name="Potassium" reversalPotential="-80mV"/> </CellEnvironment>
recording.xml
<Access id="recording">
<CurrentClamp at="p0" lineColor="red" hold="0.08nA">
</CurrentClamp>
<VoltageRecorder at="p1" lineColor="blue"/>
</Access>
na1.xml
<KSChannel id="na1" gSingle="30pS" permeantIon="Na"> <ClosedState id="c1"/> <OpenState id="o1"/> <ClosedState id="c2"/> <VHalfTransition from="c1" to="o1" vHalf = "-35mV" z="4.5e" gamma="0.8" tau="0.15ms" tauMin="0.001ms"/> <VHalfTransition from="o1" to="c2" vHalf = "-70mV" z="1.1e" gamma="0.90" tau="4.0ms" tauMin="0.01ms"/> </KSChannel>
k1.xml
<KSChannel id="k1" gSingle="30pS" permeantIon="K"> <ClosedState id="c1"/> <OpenState id="o1"/> <VHalfTransition from="c1" to="o1" vHalf = "0mV" z="1.5e" gamma="0.75" tau="3.2ms" tauMin="0.3ms"/> </KSChannel>